SeqCode: genomeDistribution
General description
Calculation of the distribution of peaks into different genome gene features.
> bin/genomeDistribution -h SeqCode_v1.0 User commands genomeDistribution NAME genomeDistribution - a program to examine the distribution of peaks within distinct genomic features. SYNOPSIS genomeDistribution [-1/7][-d][-g ][-p 1234][-q 12][-s ][-w ][-v][-x prefix][-h] OUTPUT One folder with 3 files: - Peaks associated to genomic regions - R script commands - Graphical representation (R is required). OPTIONS -1 : Color 1, Distal/Promoter (default: coral). -2 : Color 2, Proximal/Intragenic (default: darkred). -3 : Color 3, 5'UTR/Intergenic (default: burlywood). -4 : Color 4, 3'UTR (default: orchid). -5 : Color 5, CDS (default: chartreuse3). -6 : Color 6, Introns (default: gold). -7 : Color 7, Intergenic (default: corflowerblue). -d : Detailed analysis with introns/exons/promoters. -g : Gradient palette from Color1 to white. -p : Viridis palettes (1=viridis|2=magma|3=plasma|4=inferno). -q : ColorBrewer palettes (1=accent|2=paired). -s : Simplified with genic/intragenic/intergenic (X bp around TSS). -w : Window resolution (default: 100). -v : Verbose. Display info messages. -x : Prefix for the output folder. -h : Show this help. SEE ALSO SeqCode homepage: http://ldicrocelab.crg.es GitHub source code: https://github.com/eblancoga/seqcode AUTHORS Written by Enrique Blanco. SeqCode_v1.0 User commands genomeDistribution
Examples
Example 1. How to generate a comprehensive pie chart with the full set of genomic features.
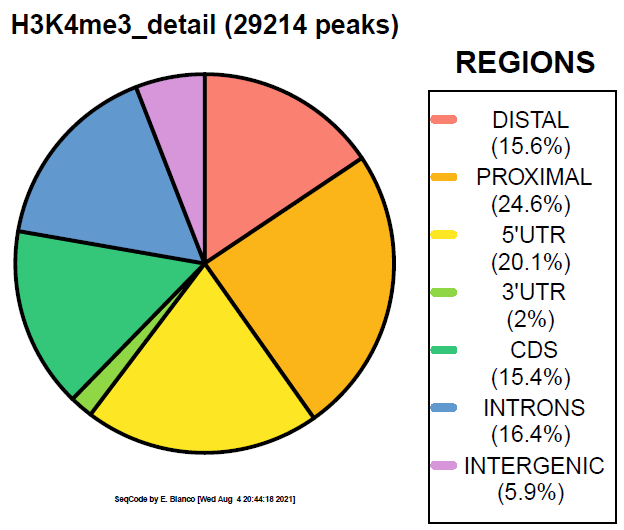

Example 2. How to colorize the piechart with the ColorBrewer accent palette.
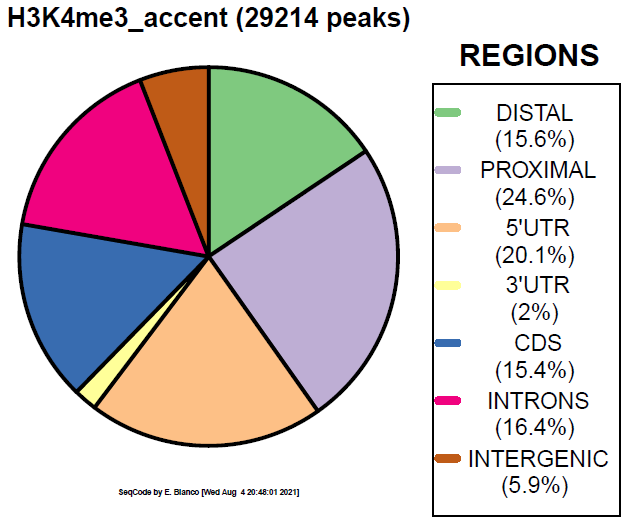

Example 3. How to colorize the piechart with the ColorBrewer paired palette.

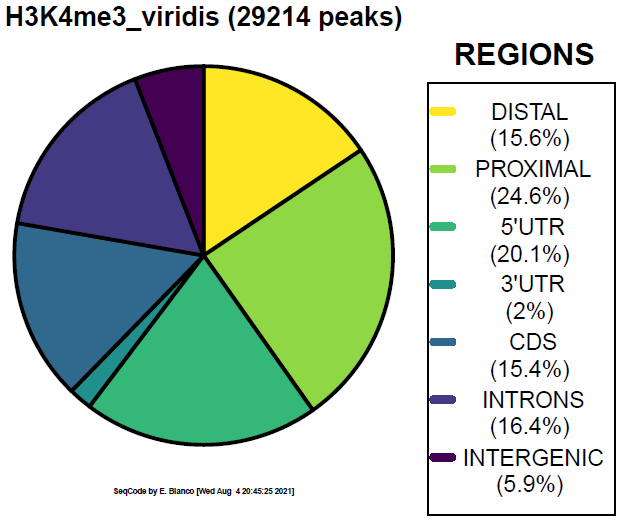
Example 4. How to colorize the piechart with the Viridis palette.

Example 5. How to colorize the piechart with the Viridis magma palette.


Example 6. How to colorize the piechart with the Viridis plasma palette.


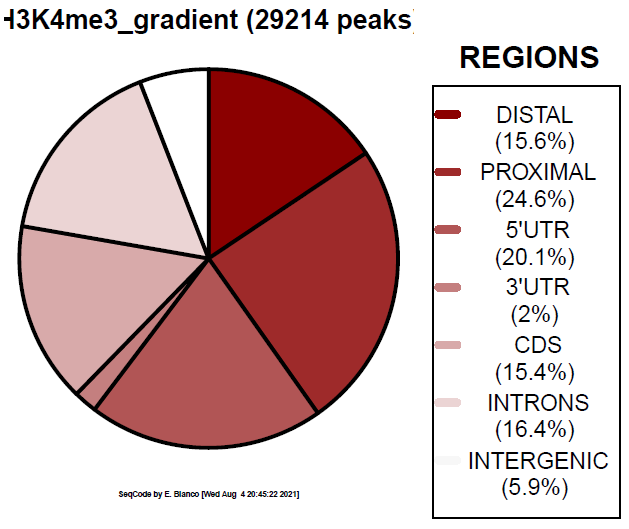
Example 7. How to colorize the piechart with the Viridis inferno palette.
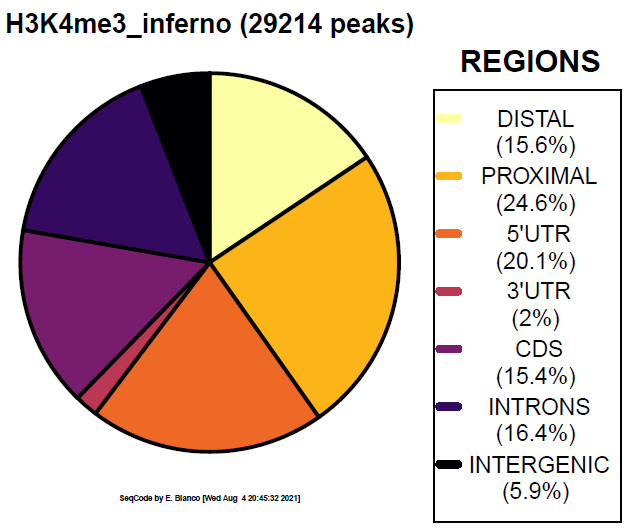

Example 8. How to colorize the piechart with the colors selected by the user.
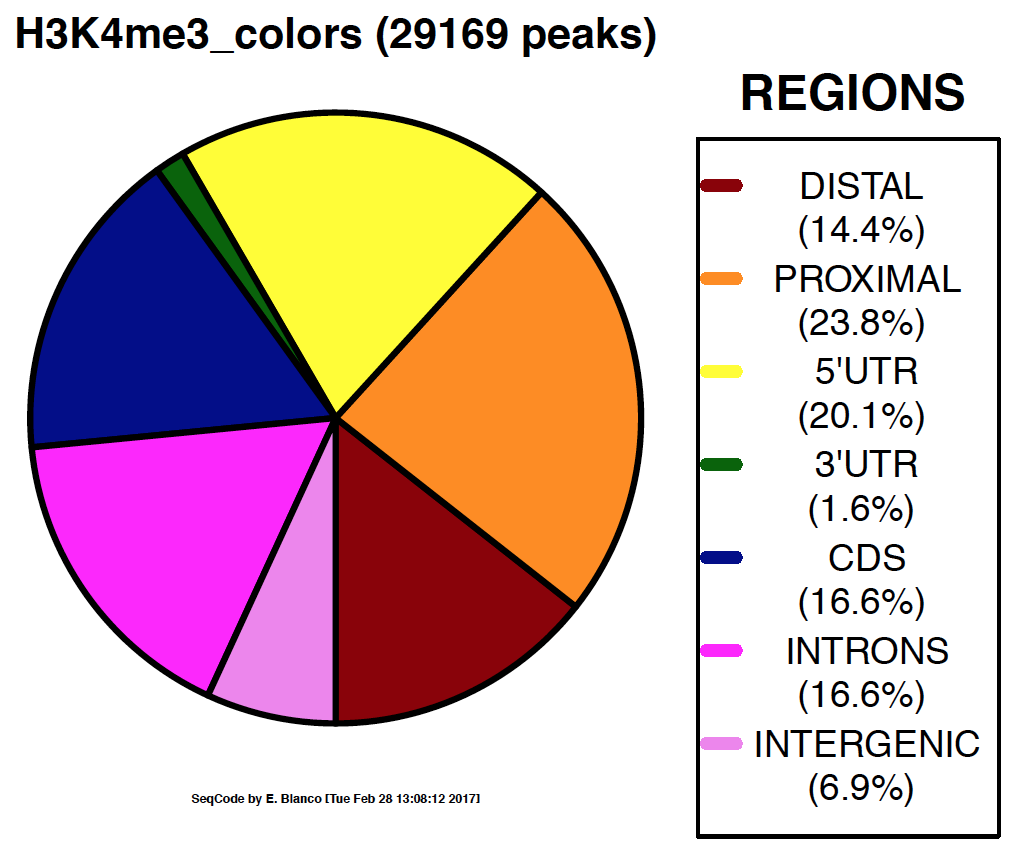

Example 9. How to colorize the piechart with a gradient of colors.

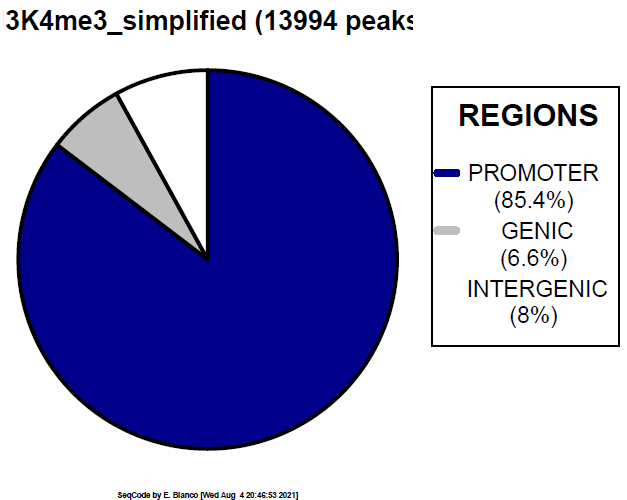
Example 10. How to generate a simplified piechart focusing on only three classes customized by distance to TSS.

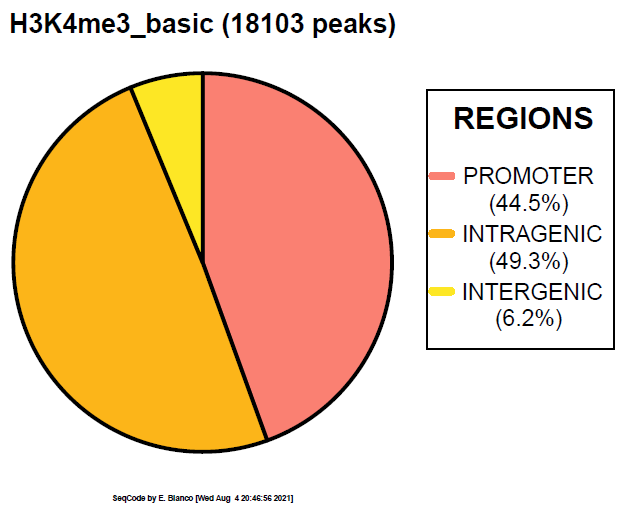
Example 11. How to generate a simple pie chart with the basic gene features (not defined by distance to TSS).

Please, note that peaks overlapping more than one genomic feature are proportionally counted the same number of times. The original number of peaks and the resulting number of peaks after the application of this rule are included into the caption of resulting plots.
> bin/genomeDistribution -vd ChromInfo.txt refGene.txt H3K4me3_peaks.bed H3K4me3_detail


Example 2. How to colorize the piechart with the ColorBrewer accent palette.
> bin/genomeDistribution -vdq 1 ChromInfo.txt refGene.txt H3K4me3_peaks.bed H3K4me3_accent


Example 3. How to colorize the piechart with the ColorBrewer paired palette.
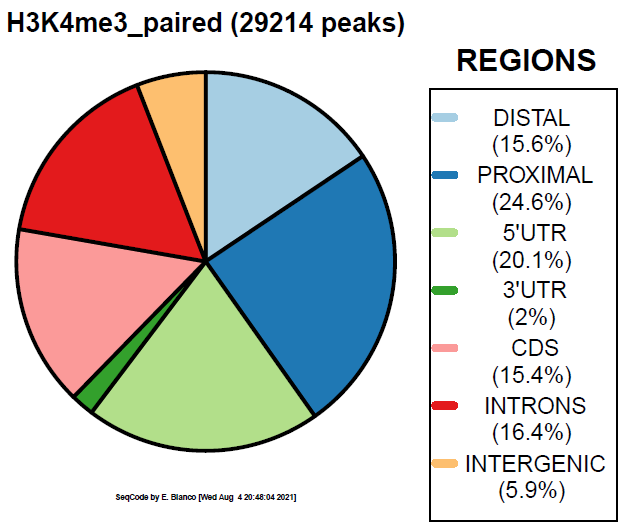
> bin/genomeDistribution -vdq 2 ChromInfo.txt refGene.txt H3K4me3_peaks.bed H3K4me3_paired


Example 4. How to colorize the piechart with the Viridis palette.
> bin/genomeDistribution -vdp 1 ChromInfo.txt refGene.txt H3K4me3_peaks.bed H3K4me3_viridis


Example 5. How to colorize the piechart with the Viridis magma palette.
> bin/genomeDistribution -vdp 2 ChromInfo.txt refGene.txt H3K4me3_peaks.bed H3K4me3_magma


Example 6. How to colorize the piechart with the Viridis plasma palette.
> bin/genomeDistribution -vdp 3 ChromInfo.txt refGene.txt H3K4me3_peaks.bed H3K4me3_plasma


Example 7. How to colorize the piechart with the Viridis inferno palette.
> bin/genomeDistribution -vdp 4 ChromInfo.txt refGene.txt H3K4me3_peaks.bed H3K4me3_inferno


Example 8. How to colorize the piechart with the colors selected by the user.
> bin/genomeDistribution -vd -1 darkred -2 darkorange -3 yellow -4 darkgreen -5 darkblue -6 magenta -7 violet ChromInfo.txt refGene.txt H3K4me3_peaks.bed H3K4me3_customized


Example 9. How to colorize the piechart with a gradient of colors.
> bin/genomeDistribution -vd -g -1 darkred $MOUSE9/ChromInfo.txt $MOUSE9/refGene.txt H3K4me3_peaks.bed H3K4me3_gradient


Example 10. How to generate a simplified piechart focusing on only three classes customized by distance to TSS.
> bin/genomeDistribution -vs 2500 -1 darkblue -2 grey -3 white ChromInfo.txt refGene.txt H3K4me3_peaks.bed H3K4me3_simplified


Example 11. How to generate a simple pie chart with the basic gene features (not defined by distance to TSS).
> bin/genomeDistribution -v ChromInfo.txt refGene.txt H3K4me3_peaks.bed H3K4me3_basic


Please, note that peaks overlapping more than one genomic feature are proportionally counted the same number of times. The original number of peaks and the resulting number of peaks after the application of this rule are included into the caption of resulting plots.
ChIPseq and RNAseq samples
Please, follow the link below for further information on how to get and preprocess the raw data of
published ChIPseq and RNAseq samples utilized in this glossary of SeqCode functions.
[SAMPLES]
[SAMPLES]